3. Examples¶
3.1. G2/M cell cycle transition¶
The SBML2Julia GitHub repository contains a version of the Vinod et Novak model of the G2/M cell cycle transition. The model contains 13 species, 13 simulated observables, 24 parameters and 4 experimental conditions.
3.1.1. Using the Python API¶
The SBML2Julia problem can be created using the Python API (and assuming that the current working directory is the SBML2Julia root directory) via:
>>> import sbml2julia
>>> problem = sbml2julia.SBML2JuliaProblem('examples/Vinod_FEBS2015/Vinod_FEBS2015.yaml')
and solved by:
>>> problem.optimize()
The results are then available under problem.results, which returns a dictionary containing 'par_best' (the best found parameter set), 'species', 'observables', 'fval' (the negative log-likelihood) and 'chi2' (chi2 values of residuals). For example, to access the best parameter set, type:
>>> problem.results['x_best']
Name par_0 par_best par_best_to_par_0
0 kDpEnsa 0.0500 0.048840 0.976807
1 kPhGw 1.0000 0.955727 0.955727
2 kDpGw1 0.2500 0.236274 0.945095
3 kDpGw2 10.0000 9.727009 0.972701
4 kWee1 0.0100 0.009697 0.969738
5 kWee2 0.9900 1.308203 1.321418
6 kPhWee 1.0000 1.064760 1.064760
7 kDpWee 10.0000 10.181804 1.018180
8 kCdc25_1 0.1000 0.135812 1.358117
9 kCdc25_2 0.9000 1.099638 1.221820
10 kPhCdc25 1.0000 0.961436 0.961436
11 kDpCdc25 10.0000 8.990541 0.899054
12 kDipEB55 0.0068 0.003400 0.500002
13 kAspEB55 57.0000 46.386552 0.813799
14 fCb 2.0000 1.999269 0.999634
15 jiWee 0.1000 0.199999 1.999988
16 fB55_wt 1.0000 0.967920 0.967920
17 fB55_iWee 0.9000 0.886548 0.985053
18 fB55_Cb_low 1.1000 1.066191 0.969265
19 fB55_pGw_weak 1.0000 0.971742 0.971742
20 kPhEnsa_wt 0.1000 0.100393 1.003933
21 kPhEnsa_iWee 0.1000 0.097819 0.978187
22 kPhEnsa_Cb_low 0.1000 0.101325 1.013248
23 kPhEnsa_pGw_weak 0.0900 0.090801 1.008902
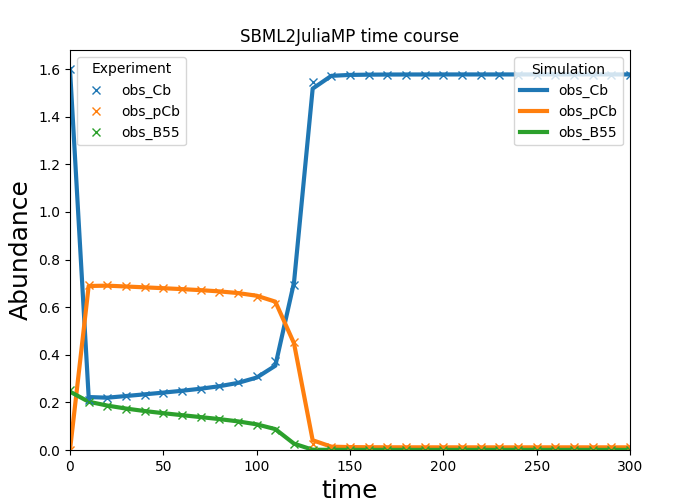
Selected observables of the optimized simulation of a given simulation condition can be plotted via:
>>> problem.plot_results(condition='wt', observables=['obs_Cb', 'obs_pCb', 'obs_B55'])
3.1.2. Using the command line interface¶
Similarly, the same example problem can be solved from the command line interface:
user@bash:/sbml2julia$ sbml2julia optimize 'examples/Vinod_FEBS2015/Vinod_FEBS2015.yaml' -d 'examples/Vinod_FEBS2015/results/'
The results can be found in the output directory given to the -d argument.
3.2. Gut microbial community¶
The SBML2Julia GitHub repository contains a generalised Lotka-Volterra model of 12 gut bacteria. The model conceived by Shin et al. contains 2 species, 2 observables, 156 parameters and 210 experimental conditions in its PEtab formulation.
3.2.1. Using the Python API¶
The SBML2Julia problem can be created using the Python API (and assuming that the current working directory is the sbml2julia root directory) via:
>>> import sbml2julia
>>> problem = sbml2julia.SBML2JuliaProblem('examples/Shin_PLOS2019/Shin_PLOS2019.yaml'})
and solved by:
>>> problem.optimize()
Again, the results are available under problem.results. Plots of the observables can be generated with the problem.plot_results() method and results can be written to TSV files with problem.write_results().
3.2.2. Using the command line interface¶
Similarly, the same example problem can be solved from the command line interface:
user@bash:/sbml2julia$ sbml2julia optimize 'examples/Shin_PLOS2019/Shin_PLOS2019.yaml' -d './examples/Shin_PLOS2019/results/'
The results can be found in the output directory given to the -d argument.